ML4Q Publishing Guide
ML4Q is committed to the principles of reproducibility, scientific integrity, and open access. These practices are slowly becoming community standards and are increasingly demanded by publishers. Additionally, the process of preparing data and analysis for other eyes is an additional step that can improve the quality and reliability of the work. It also will enable the use of data by other groups, thus creating new possibilities and synergies for the scientific community and beyond. This document tries to provide a guide you can follow along, when preparing your research data for publication.

Figure 1: 3 steps for publishing research data
The guide is divided into 3 steps. For preparing the research data, there are multiple possibilities categorized into must-haves, strong recommendations, and nice-to-haves. Depending on the type of publication (experimental, theoretical, or numerical), some may not apply to you. Publication is shown exemplary with zenodo.org. We recommend publishing data on zenodo.org or on the journal’s own data repository. Finally, after publication, the DOI of the data is referenced in the publication and be independently cited and shared.
Preparation of research data
To make research data available, you will usually be uploading it to a data repository as a single folder. The data you will upload will be openly accessible, and it will stay that way indefinitely. Therefore, it is necessary that you prepare your data folder for publication. This requires time and effort, and for every project the requirements are different.
There are different relevant levels of publishing data:
- Minimal data publication: Publication of the numerical data shown in figures in a format that is readable by others.
- Representative data publication: Publication of representative raw/processed data, depending on data and situation.
- Full data publication: Publication of the raw data and scripts for the full data processing chain that produce the final plots shown in the paper.
For ML4Q funded publications, the minimal data publication is mandatory.
Must-haves
- Structure your data folder in a logical and understandable manner. Prevent deep folder hierarchies, as well as putting every file into the root folder. Instead, the root folder could contain a readme file (see below) and subfolders, which then contain the files. For example, you could have separate subfolders for raw data, processed data, code/scripts, and figures.
- Create a legible readme file in the folder that describes what the data is, where to find which parts of the data, and (if applicable) what needs to be done to reproduce the results. It is best if someone who does not know the project can understand the entire project based on the readme – this also includes yourself in a few years from now!
- Include at least the data you use in your publication. This means, that every figure in your publication should be accompanied by the extracted data in a format that is readable to others. Ideally, you should aim to export your data in a common format, for example csv files or hdf5 files. Binary representation of data can save space but can make the data harder to access. Avoid data formats that need proprietary software to view. It is acceptable to upload the data in another format, as long as that data file is accompanied by an instruction on how to load the data. For example, one could include a piece of matlab or python code that would load the data for the reader. As long as it is documented properly how to load the data, this is sufficient.
- Make sure no privacy-sensitive information is included in the data. Remove non-shareable data objects (raw and processed!), passwords hardcoded in your scripts, comments containing private information, and so on.
Recommendations
- Include your raw and processed data that serve as input for the publication figures. This does not mean, that every data ever recorded should be included. If some data is not used for some reason, leave it out. However, it is good practice to mention if and how data was selected.
- If you only include representative data, that is already processed from your raw data in some way, document, how that data was collected/processed.
- Include your source code and/or scripts you used to process the data and create your results / output the figures. Record the software packages that you used, including their versions. Optimally, people should be able to reproduce your results on their own.
- If you use well maintained packages within your data analysis, e.g. when using python, you would use numpy, scipy, pandas and matplotlib for the data processing and plotting, you can specify the versions that are used
- If you want to include less established source code and/or scripts from a public code repository, include a snapshot here. Collaboration and versioning tools like github, gitlab, etc. are not suitable for publication, since the versioning history can be rewritten, projects may be deleted or moved, and URLs can change over time. It is good practice to cite the code you use.
Nice-to-haves
- Reformat the code so that it is portable and easily reproducible. This means that when someone else downloads the data, they do not need to change the code to run it. For example, this means that you do not read data with absolute paths (e.g., C:/my_name/Documents/PhD/projects/project_title/raw_data/measurement.hd5) on your computer, but only to relative paths on the project (e.g., raw_data/measurement.hd5).
- In principle Zenodo DOI can be linked to binder to host related ipython notebooks online, which makes it more accessible (https://blog.jupyter.org/binder-with-zenodo-af68ed6648a6)
- Format your code so that it is legible by others. Write informative comments, split up your scripts in logical chunks, and use a consistent style.
- If you have privacy-sensitive data, it may still be possible to create example data for others to run the code on. This ensures maximum reproducibility.
Publication
Data Repositories
When choosing the appropriate data repository for the data publication, you have different possibilities:
- Zenodo
- Journal repositories
- Institutional repositories
The most general and often the most sensible choice is Zenodo. Zenodo is a general-purpose open repository developed under the European OpenAIRE program and operated by CERN. It is used by many scientists for (data) publication. Data publications on Zenodo can be grouped together, for example for all ML4Q publications, and are searchable and findable by others.
Nowadays, many journals operate their own data repository. These are also good choices for data publications, that accompany a text publication.
If your university or research institute offers a data repository, it may also be suitable for data publication, if it satisfies your requirements.
Publication on Zenodo
Using Zenodo as an example, the data publication process is described below, including important metadata, choice of license, etc.
Compress your data folder
Compress the complete data folder into a single archive file. It is recommended to use zip, as it is supported by practically every operating system natively. After compressing, check the contents for completeness and unnecessary files. Your data is now ready to be published.
Create an account
To upload anything to Zenodo, you need an account. If you already have an ORCID or a GitHub account, then you can link these directly to your Zenodo login and login through these.
Go to https://zenodo.org/ and click on Sign up. Follow the steps and complete the account creation. Afterwards, login.
Upload to data repository
In the upper right corner, you find a button to create a new upload.
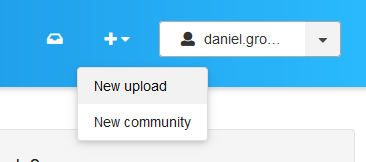
Figure 2: New upload at Zenodo
When you click the upload button, you will get a page where you can upload your files, determine the type of upload, and create metadata for the research object. You can just drag and drop your zip archive to the upload window.
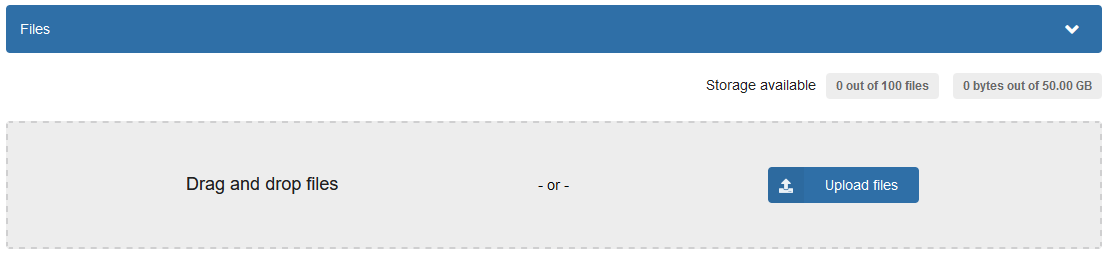
Figure 3: Upload data dialog
A Zenodo publication can be linked to Zenodo communities to bundle publications together and simplify finding similar publications. Please add your ML4Q funded publication at least to the ML4Q community. Since a publication can be linked to multiple communities, you can of course add other communities as you see fit. You find the button to select a community on top of the site, above the file upload window.
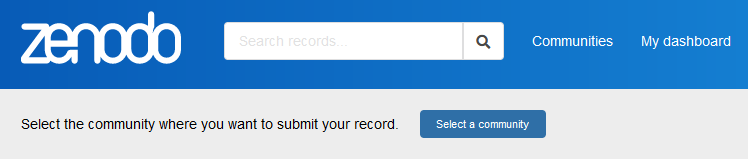
Figure 4: Add the publication to one or more communities
Below the upload window is the dataset metadata. Fill out the basic information. For resource type, you may choose e.g. dataset. Choose a meaningful title, for example Data and code for “title of publication”. Make sure to add all authors and affiliations. The description should contain information about what the data is and where to find which parts of the data. You can use the content of your readme file.
Make sure to choose the correct license for your data. If the data should be openly re-used and re-distributed, the default Creative Commons Attribution 4.0 International is a good choice as data license. For details about the license, see https://creativecommons.org/licenses/by/4.0/.
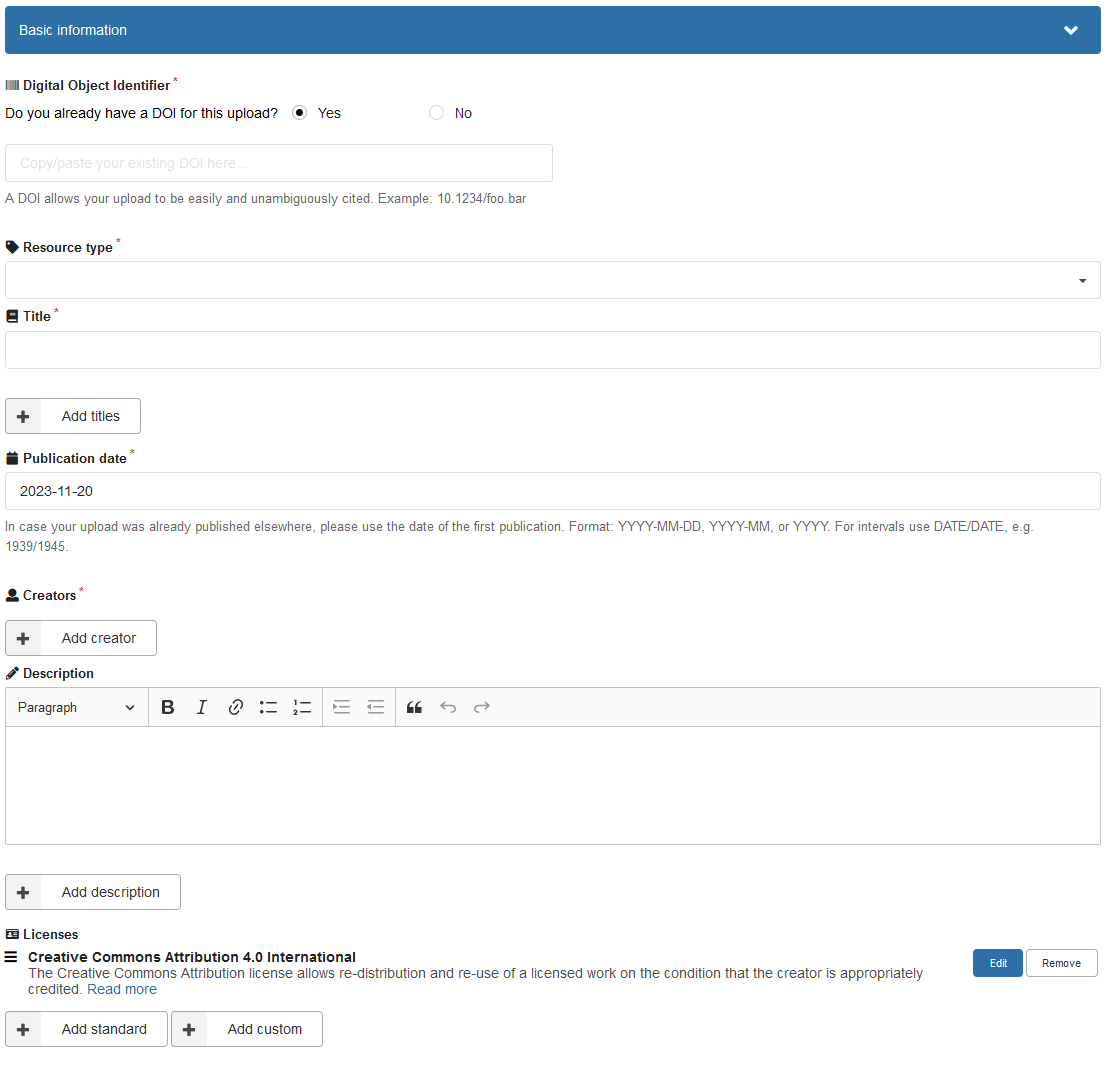
Figure 5: Basic information entry
Below the basic information you can enter additional metadata. These include among other things contributors (differentiated from authors/creators), languages, dataset version, funding information, related works and references and information about the text publication.
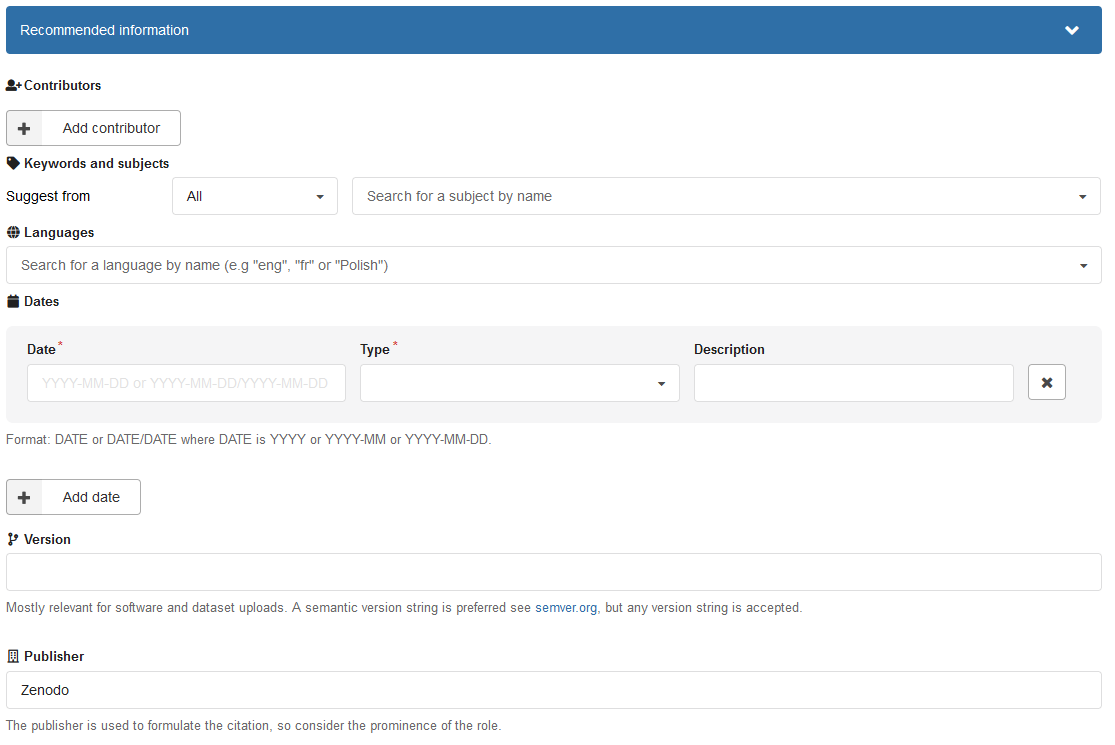
Figure 6: Recommended information entry
The last step is to save the draft, preview and publish. After publishing, you get a DOI to the published data. Your research code is now findable, citable, understandable, reproducible, and archived. If you find errors and you want to change the data, you can also upload a new version of the same project on Zenodo.
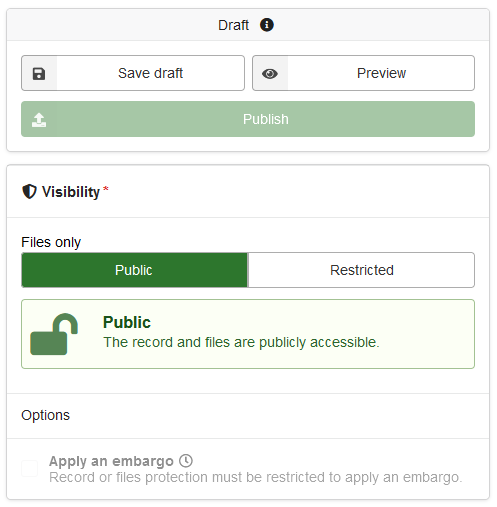
Figure 7: Dataset settings
Citation
You can now add the DOI of the research data to your text publication. For example, add a paragraph to your Acknowledgements chapter:
Data and materials availability: Raw data as well as all measurement, data-analysis, and simulation code used in the generation of main and supplementary figures are available in Zenodo with the identifier 10.1234/zenodo.xyz.
In addition, add the DOI to your data publication to the metadata of your text publication.
References
Akhmerov, A., & Steele, G. (2019). Open Data Policy of the Quantum Nanoscience Department, TU Delft. Zenodo. https://doi.org/10.5281/zenodo.2556949
Kesteren, E.-J. (2017). A short practical guide for preparing and sharing your analysis code. https://odissei-data.nl/en/2022/06/a-short-practical-guide-for-preparing-and-sharing-your-analysis-code/